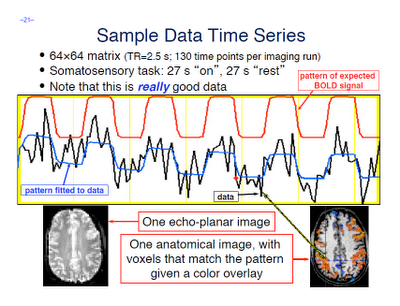
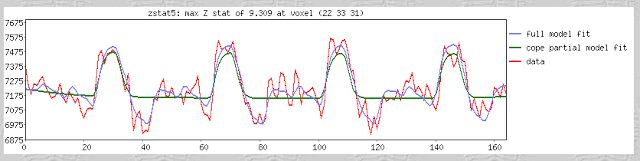
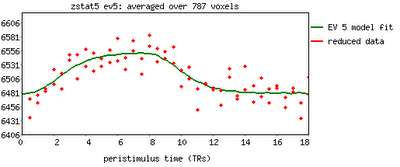
As promised, we now begin our series of AFNI tutorials. These walkthroughs will be more in-depth than the FSL series, as I am more familiar with AFNI and use it for a greater number of tasks; accordingly, more advanced tools and concepts will be covered.
Using AFNI requires a solid understanding of Unix; the user should know how to write and read conditional statements and for loops, as well as know how to interpret scripts written by others. Furthermore, when confronted with a new or unfamiliar script or command, the user should be able to make an educated guess about what it does. AFNI also demands a sophisticated knowledge of fMRI preprocessing steps and statistical analysis, as AFNI allows the user more opportunity to customize his script.
A few other points about AFNI:
1) There is no release schedule. This means that there is no fixed date for the release of new versions or patches; rather, AFNI responds to user demands on an ad hoc basis. In a sense, all users are beta testers for life. The advantage is that requests are addressed quickly; I once made a feature request at an AFNI bootcamp, and the developers updated the software before I returned home the following week.
2) AFNI is almost entirely run from the command line. In order to make the process less painful, the developers have created "uber" scripts which allow the user to input experiment information through a graphical user interface and generate a preprocessing script. However, these should be treated as templates subject to further alteration.
3) AFNI has a quirky, strange, and, at times, shocking sense of humor. Through clicking on a random hotspot on the AFNI interface, one can choose their favorite Shakespeare sonnet; read through the Declaration of Independence; generate an inspirational quote or receive kind and thoughtful parting words. Do not let this deter you. As you become more proficient with AFNI, and as you gain greater life experience and maturity, the style of the software will become more comprehensible, even enjoyable. It is said that one knows he going insane when what used to be nonsensical gibberish starts to take on profound meaning. So too with AFNI.
The next video will cover the to3d command and the conversion of raw volumetric data into AFNI's BRIK/HEAD format; study this alongside data conversion through mricron, as both produce a similar result and can be used to complement each other. As we progress, we will methodically work through the preprocessing stream and how to visualize the output with AFNI, with an emphasis on detecting artifacts and understanding what is being done at each step. Along the way different AFNI tools and scripts will be broken down and discussed.
At long last my children, we shall take that which is rightfully ours. We shall become as gods among fMRI researchers - wise as serpents, harmless as doves. Seek to understand AFNI with an open heart, and I will gather you unto my terrible and innumerable flesh and hasten your annihilation.